Introduction
About this document.
This is a living document, as the Ebola epidemic is rapidly evolving. Presented below is a preliminary modeling approach to the crisis, originally conceived as an illustration of the spatial SEIR model family generally and the capabilities of the rapidly developing (and totally unfinished) libspatialSEIR software library particularly. This is not yet peer reviewed research, or even a particularly complete analysis. Nevertheless, we hope that the initial exploration given below is instructive and useful.
Past versions of this analysis are cached in a repository on Github, and remain available:
- Sep 8, 2014
- Aug 28, 2014
- Aug 12, 2014
- Aug 10, 2014
- Aug 5, 2014
- Aug 3, 2014
- Jul 30, 2014
- Jul 29, 2014
- Jul 25, 2014
- Jul 25, 2014
Additional analyses are underway. An informal look at prediction performance over time is also available.
Summary of Recent Changes
- Oct 2 There have been a lot of changes for this release. The temporal component was dramatically simplified, so we're fitting a baseline SEIR model with a different intensity parameter for each nation. In addition, the spatial structure was generalized to allow a separate mixing parameter for each pair of countries. Finally, three different methods of calculating R0 are compared, and a lot of behind the scenes changes to libspatialSEIR and the R API took place. Upcoming changes include the introduction of "hybrid" samplers which may help deal with some of the autocorrelation, seen for example in the E to I transition probability chains. Despite the model changes, we continue to see a dramatic and prolonged predicted increase in cases.
- Sep 8 Fixed R0 calculations, which were showing the per-report R0 components, but didn't account for infection length. They have now been appropriately integrated over time.
- Sep 4 The situation continues to worsen, which is especially worrying given that the data used are almost certainly undercounts. Removed the polynomial analysis in favor of the spline basis analysis to save on computation time.
- Aug 31 Basic reproductive number clearly very heterogenous between countries. Other interpretations TBD.
- Aug 28 With the addition of Nigeria and new data, the situation appears dire across the board, especially in Liberia. The basic reproductive number calculations no longer look reasonable, so more must be done to estimate them separately for each country. The changes may also be due to recent changes to the R0 estimation code, which must now be reevaluated. This work is underway.
- Aug 12 Liberia continues to worsen, though Sierra Leone appears to have leveled off. Guinea might be worsening slightly.
- Aug 10 Predictions remain much the same, though perhaps not so immediately catestrophic as the August 5 predictions.
- Aug 5 Models begin to show catastrophic predictions. Epidemic is changing too quickly for these simple models.
- Shorten prediction window
- Increase intensity process flexibility (quartic rather than cubic)
- Recall that these models can't account for changing inteventions, of which there are many. The predictions are therefore made under the assumption that the epidemic will continue to evolve according to the same process it has so far.
- Aug 3 Predicted epidmic curves start to strongly favor a uniformly worsening situation.
- July 30 Previous predictions of mid-fall resolution of epidemic begin to shift.
The Outbreak
The 2014 Ebola outbreak in West Africa is an ongoing public health crisis, which has killed thousands of people so far. The cross-border nature of this epidemic, which emerged in Guinea, Liberia and Sierra Leone has complicated mitigation efforts, as has the poor health infrastructure in the region. This document explores a simple spatial SEIR model to make some initial predictions.
The Data
A summary of the WHO case reports is very helpfully compiled on wikipedia. It can be easily read into R with the xml library:
With data in hand, let's begin where every analysis should begin: graphs.
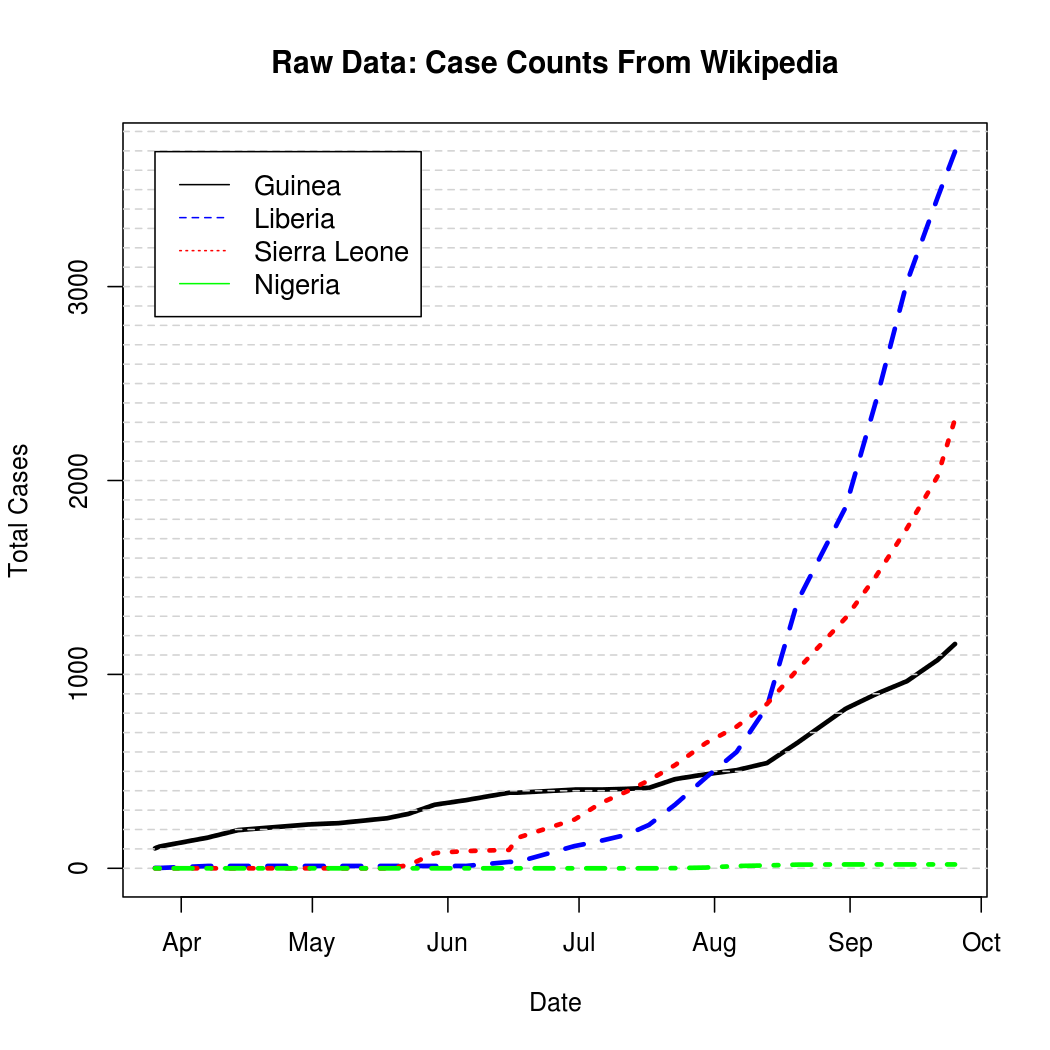
In addition to the above graph, the raw data is archived for posterity in the code block below. Wikipedia changes a lot, so it's important to record the context of the results presented here.
This graph and corresponding table represent cumulative counts, but because case reports can be revised downward due to non-Ebola illnesses the graphs are not strictly monotone. A quick, but effective solution to this problem is to simply "un-cumulate" the data and bound it at zero to get a rough estimate of new case counts over time.
For better graphical representation, the "un-cumulated" counts are scaled to represent average number of infections per day, and linearly interpolated. The process is a bit noisier from this perspective when compared to the original cumulative counts.
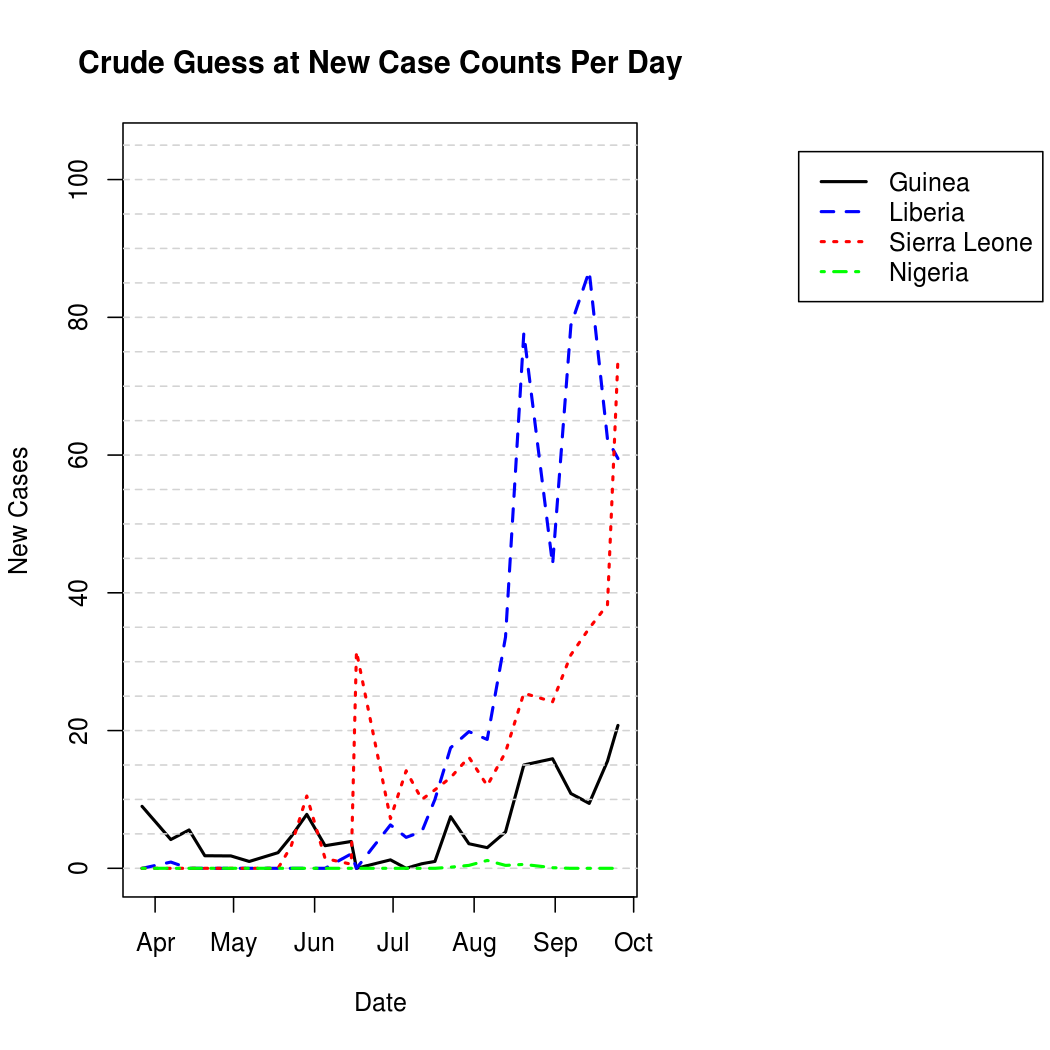
One can also represent this data geographically to get an idea of the spatial epidemic pattern, and to place the problem in a more relatable context.
Average Number of Infections Per Day:
Compartmental Models
Now that the data is read in (and now that we have several plots to suggest that we haven't done anything too terribly stupid with it) , let's do some compartmental epidemic modeling. Not only has Ebola been well modeled in the past using compartmental modeling techniques, but this author happens to be working on a software library designed to fit compartmental models in the spatial SEIRS family. What a strange coincidence! Specifically, we'll be using heirarchical Bayesian estimation methods to fit a spatial SEIR model to the data.
While a full treatment of this field of epidemic modeling is (far) beyond the scope of this writing, the basic idea is pretty intuitive. In order to come up with a simplified model of a disease process, discrete disease states (aka, compartments) are defined. The most common of these are S, E, I, and R which stand for:
- Susceptible to a particular disease
- Exposed and infected, but not yet infectious
- Infectious and capable of transmitting the disease
- Removed or recovered
This sequence, traversed by members of a population (S to E to I to R), forms what we might call the temporal process model of our analysis. This analysis belongs to the stochastic branch of the compartmental modeling family, which has its roots in deterministic systems of ordinary and partial differential equations. In the stochastic framework, transitions between the compartments occur according to unknown probabilities. It is the S to E probability, which captures infection activity, into which we introduce spatial structure. Some details of this are given as comments to the code below, and more information than you probably want on the statistical particulars is available in this pdf document. For now, suffice it to say that we'll place a simple spatial structure on the epidemic process which simply allows disease to spread between the three nations involved, and we'll try to estimate the strength of that relationship. Many other potential structures are possible, limited primarily by the amount of additional research and data compilation one is willing to do.
For the purposes of this analysis, we will not do anything fancy with demographic information or public health intervention dates. Demographic parameters are relatively difficult to estimate here, as there are only four spatial units which are all from the same region. Intervention dates are more promising, but their inclusion requires much more background research than we have time for here. In the interest of simplicity and estimability, we'll just fit a different disease intensity parameter for each of the three countries to capture aggregate differences in Ebola susceptibility in addition to using a set of basis functions to capture the temporal trend.
Analysis
Set Up
There are some things we need to define before we can start fitting models and making predictions.
- The population sizes need to be determined.
- Initial values for the four compartments must be determined.
- The time points are not evenly spaced, so we need to define appropriate offset values to capture the amount of aggregation performed (time between reports).
- We must define the spatial correlation structure.
- (optionally) A set of basis functions to capture the temporal trend.
- Prior parameters and parameter staring values must be specified for each chain.
- A whole bunch of bookkeeping stuff for which I haven't yet programmed sensible default behavior needs to be set up.
Compartment starting values follow the usual convention of letting the entire initial population be divided into susceptibles and infectious individuals. The starting value for the number of infectious individuals was 86 for Guinea and zero for the other nations. Temporal offsets are actually calculated in the first code block (above) as the differences between the report times. For temporal basis functions, several options have been explored over the life of this project, including orthogonal polynomial and natural splines. The current analysis uses no temporal basis function, instead choosing to increase the granularity of the spatial correlation structure. Future work will introduce time varying intervention terms to quantify the effects of the early efforts of MSF and the ongoing international response. Prior parameters for the E to I and I to R transitions were chosen based on well documented values for the average latent and infectious times, and the rest of the prior parameters were left vague. These decisions are addressed in more detail as comments to the code below.
With the set up out of the way, we can finally build and run the models the models. The code presented below has recently (8/27/2014) been set up to use the "parallel" library which is included with the R statistical analysis software. While this allows us to spend considerably less time waiting for the document to compile, the code may be more difficult to understand for those unfamiliar with this useful parallelization library. The previous analyses, linked above, may be a better guide to the basic use of the spatialSEIR library for such users.
Convergence
As this is a Bayesian analysis in which the posterior distribution is sampled using MCMC techniques, we really need some indication that the samplers have indeed converged to the posterior distribution in order to make any inferences about the problem at hand. In the code below, we'll read in the MCMC output files created so far, plot the three chains for each of several important parameters, and take a look at the Gelman and Rubin convergence diagnostic (which should be close to 1 if the chains have converged.)
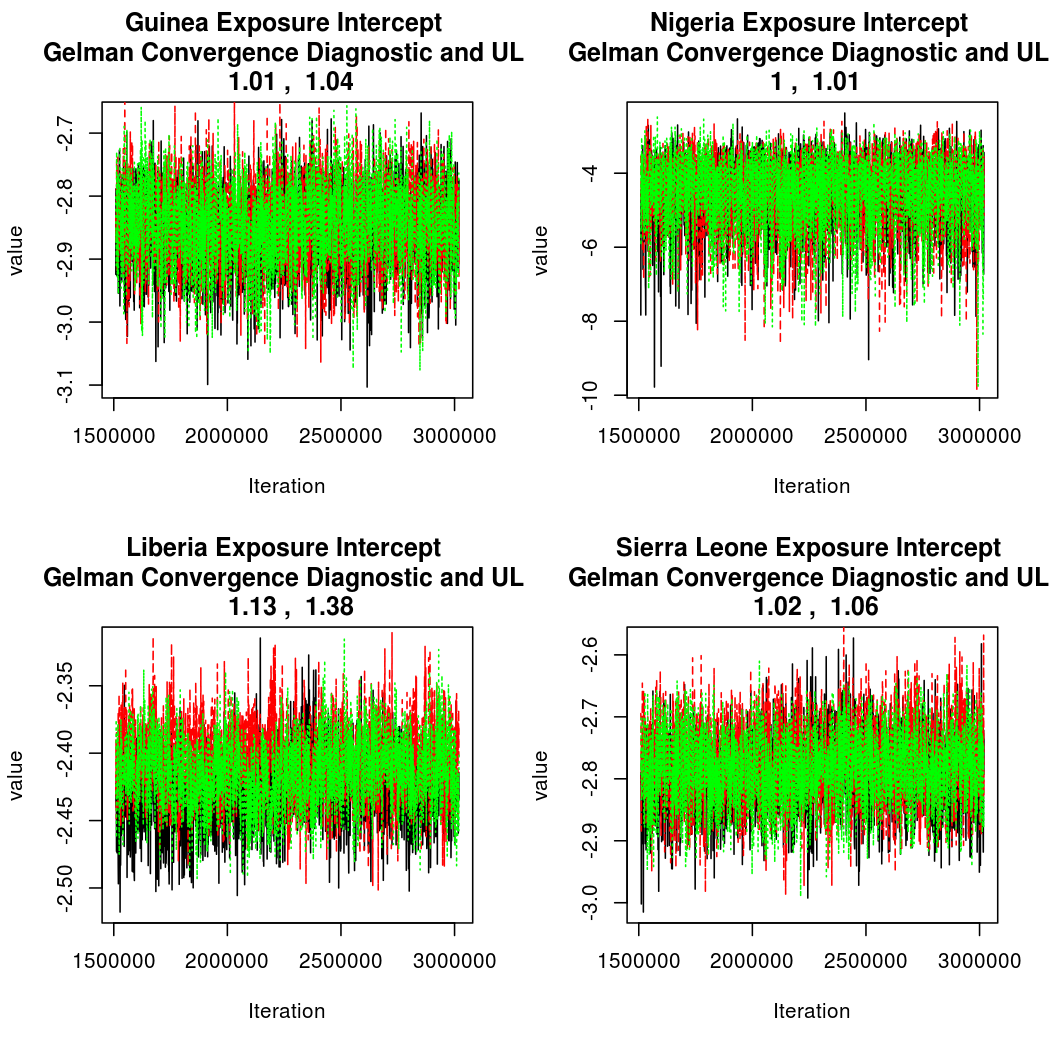
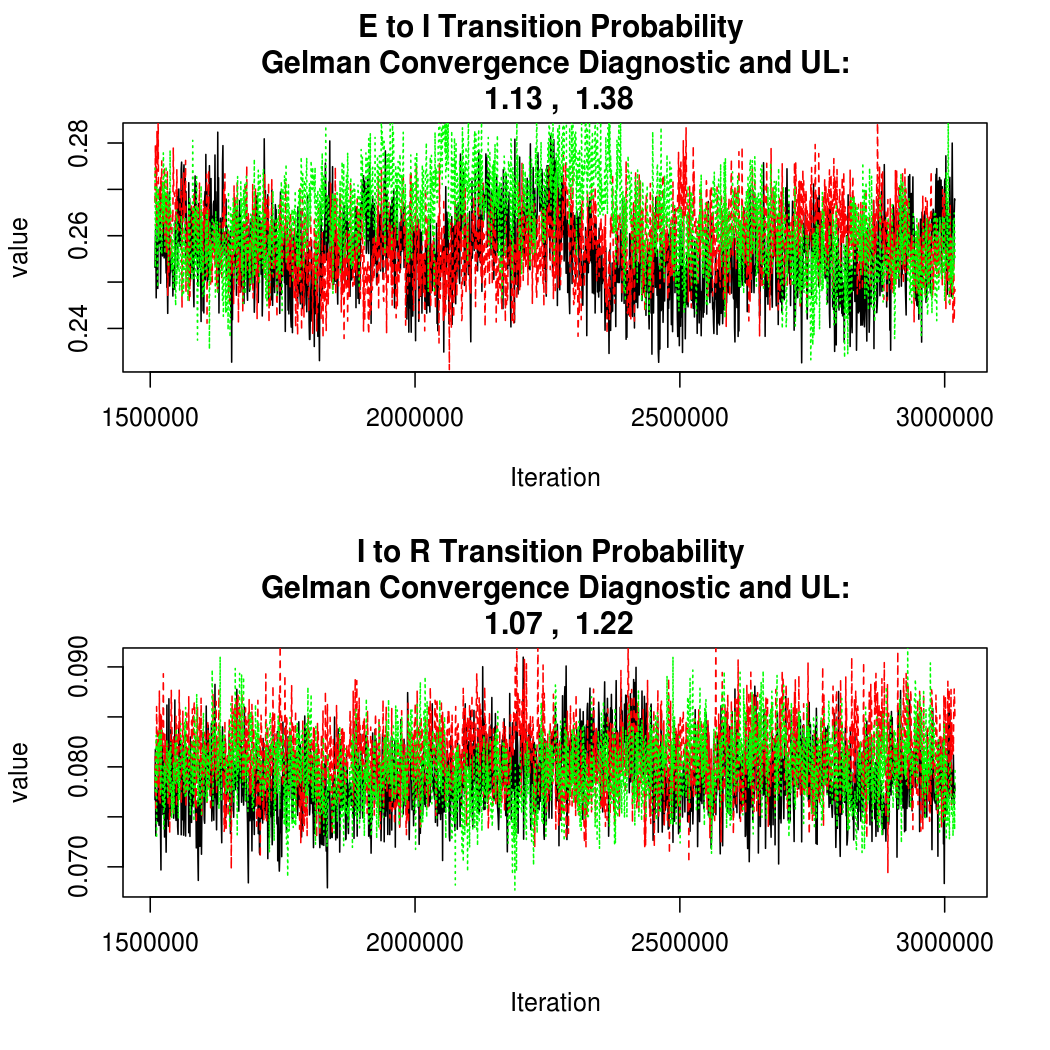
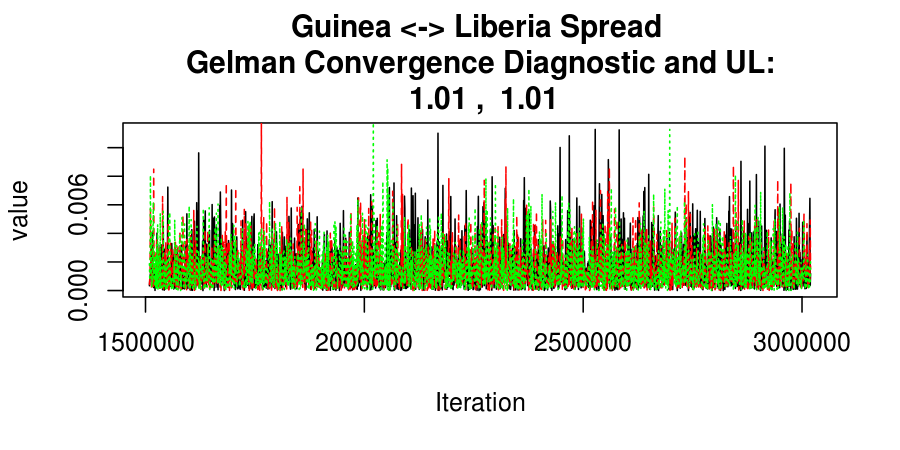
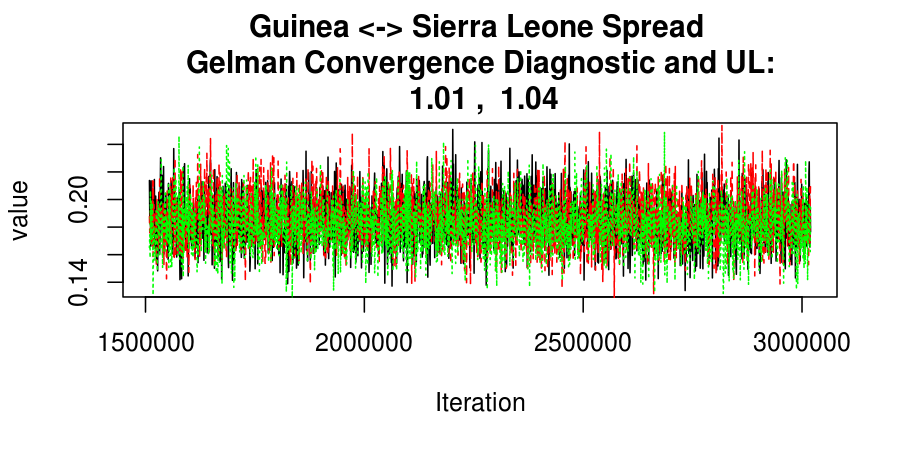
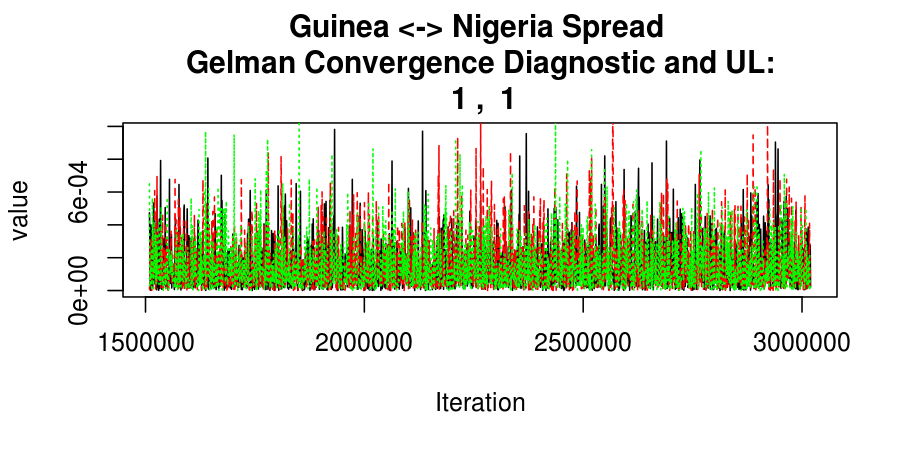
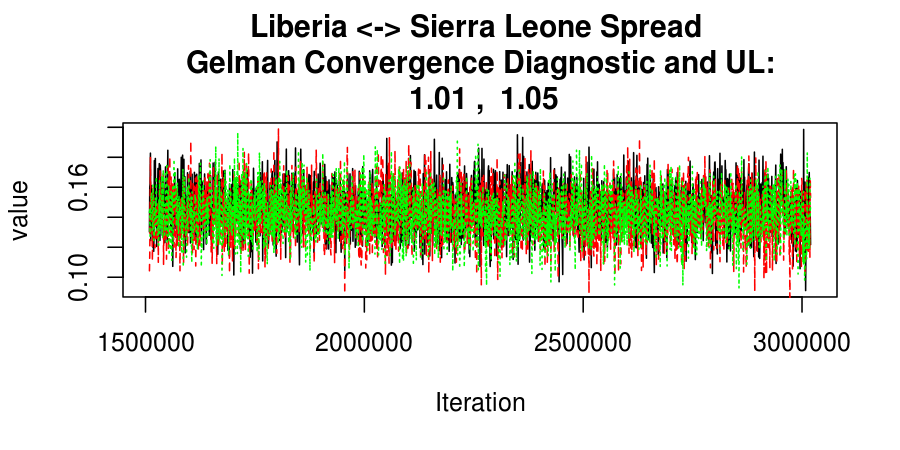
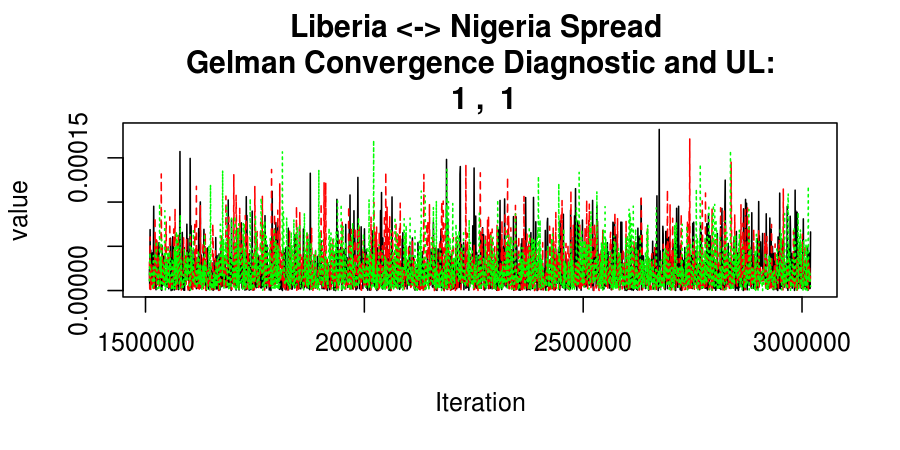
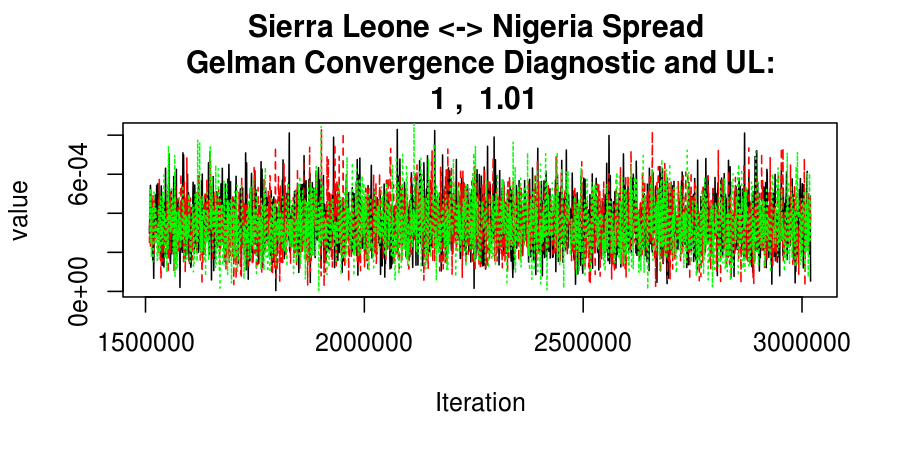
Basic Reproductive Number Calculation
A common tool for describing the evolution of an epidemic is a quantity known as the basic reproductive numer, the basic reproductive ratio, or one of several other variants on that theme. The basic idea is to quantify how many secondary infections a single infectious individual is expected to cause in a large, fully susceptible population. Naturally, when this ratio exceeds one we expect the epidemic to spread. Conversely, a basic reproductive number less than one indicates that a pathogen is more likely to die out.
In truth, there is a lot more to "basic reproductive number" calculation, especially given that different authors use different names for the same quantity, and vice versa. Here we present three different versions. First, we introduce a fairly standard "time varying basic reproductive number". This quantity is based on the intensity process parameter estimates over time, and is described in Lekone and Finkenstadt (2006). Also described in this work is the "effective" reproductive rate, which is the same quantity scaled by the number of susceptibles (almost the same in this case, due to the relatively small infectious fraction). Finally, we introduce our own, tentatively titled "empirical reproductive number". This measure follows actual transmission probabilities through time and looks at the number of secondary cases produced, on average, by one of the infectious individuals in a particular location at a particular time point. While still influenced by the parametric form chosen, this alternative approach to basic reproductive number calculation may more closely reflect population dynamics.
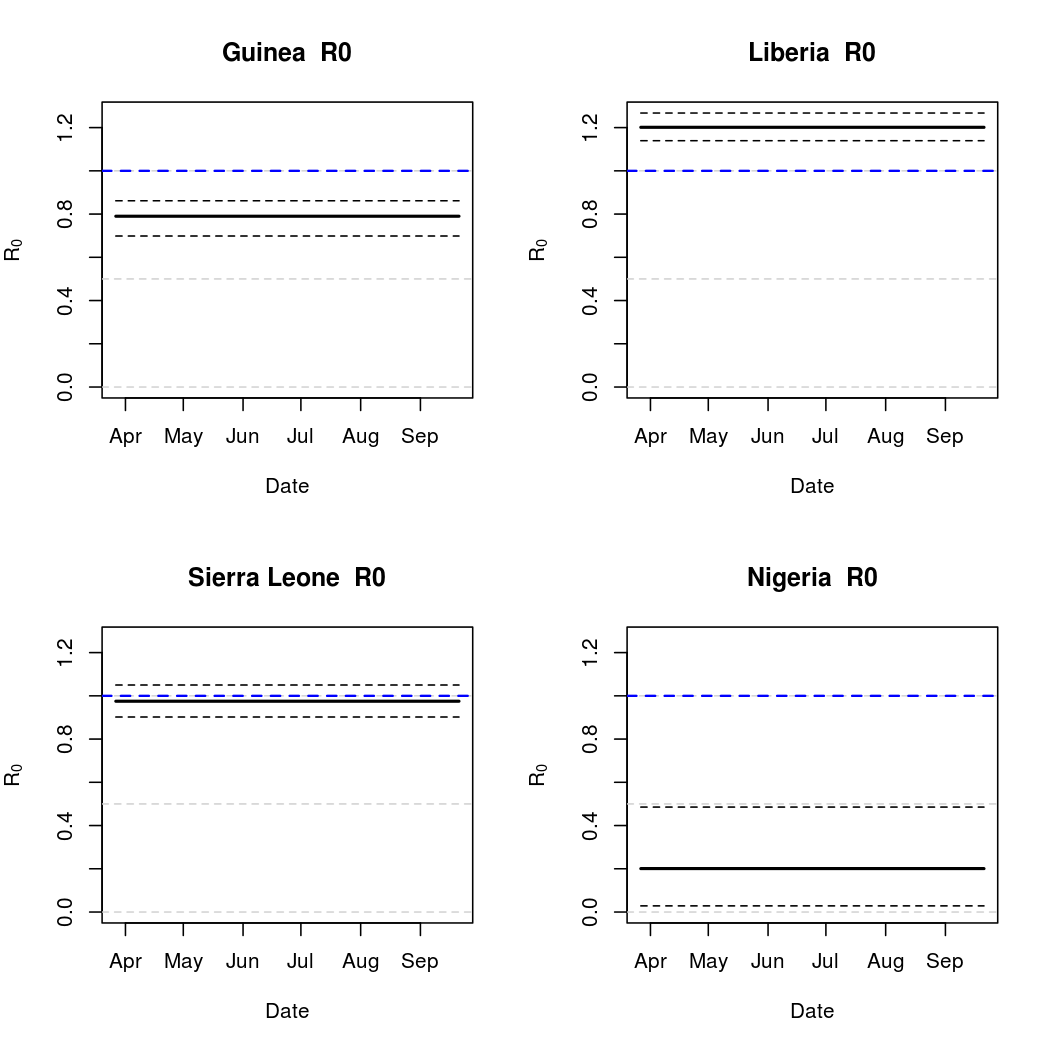
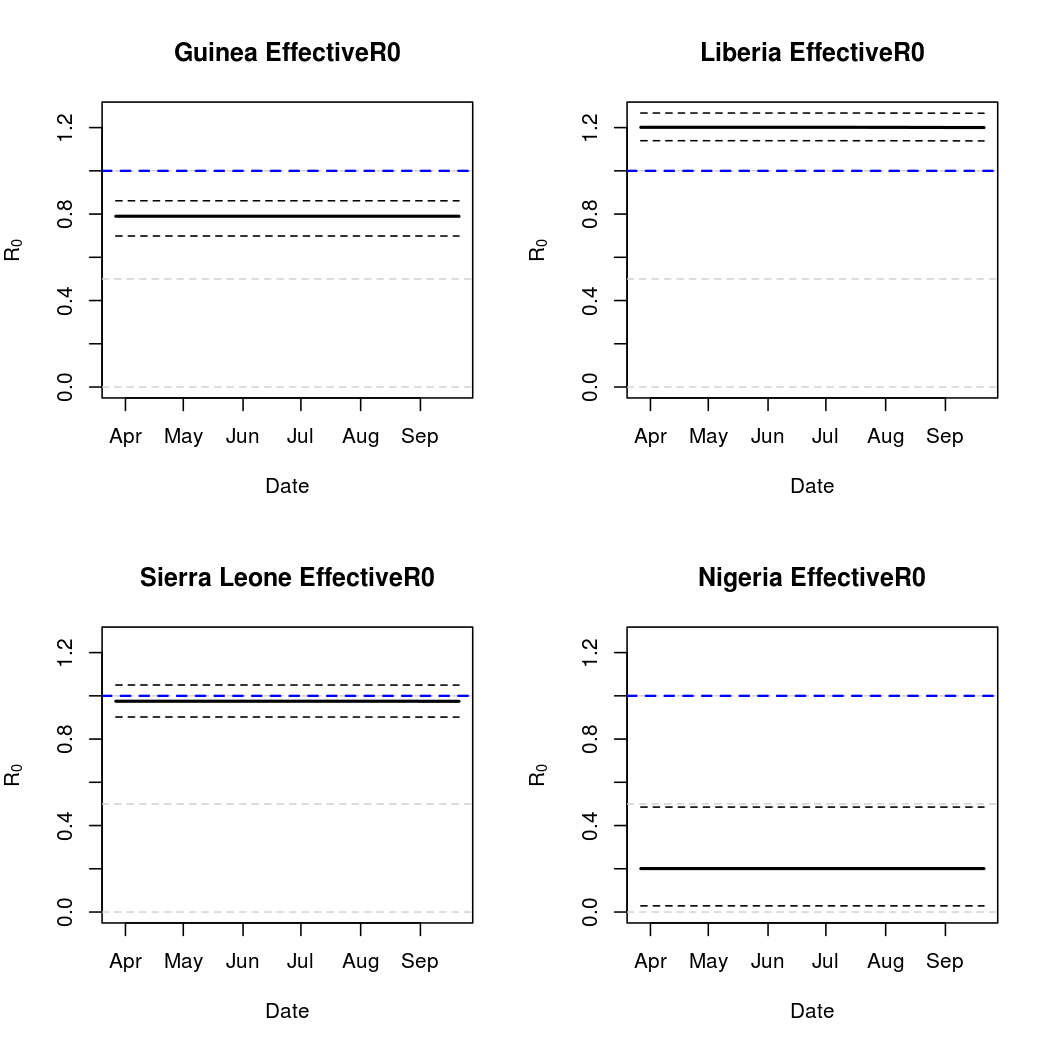
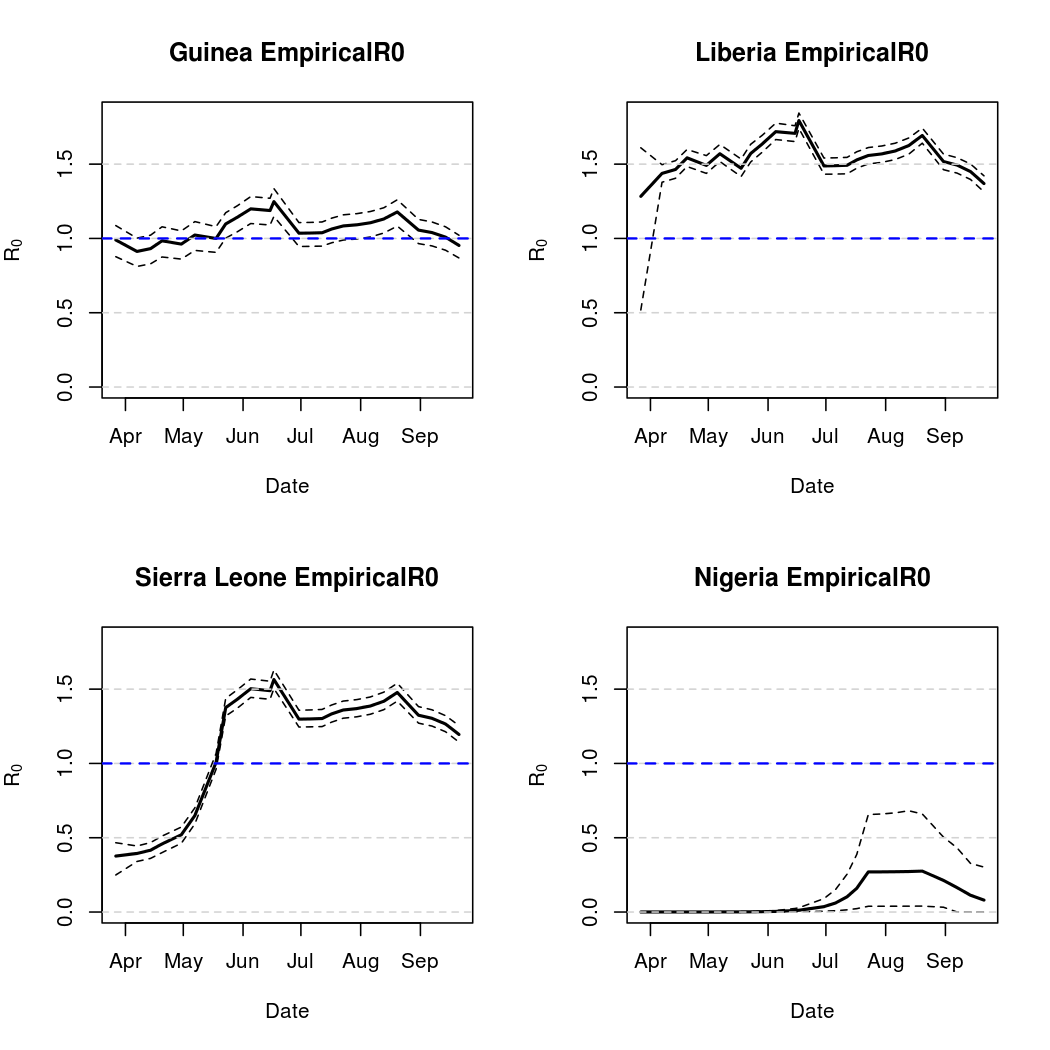
Epidemic Prediction
Below, we will attempt to predict the course of the epidemic through early fall. We must be cautious when making predictions about a chaotic process this far into the future. The intensity function which drives the exposure process is based on simple smooth functions of time, rather than any external information. While this is perfectly adequate for estimation, it may or may not provide good prediction performance.
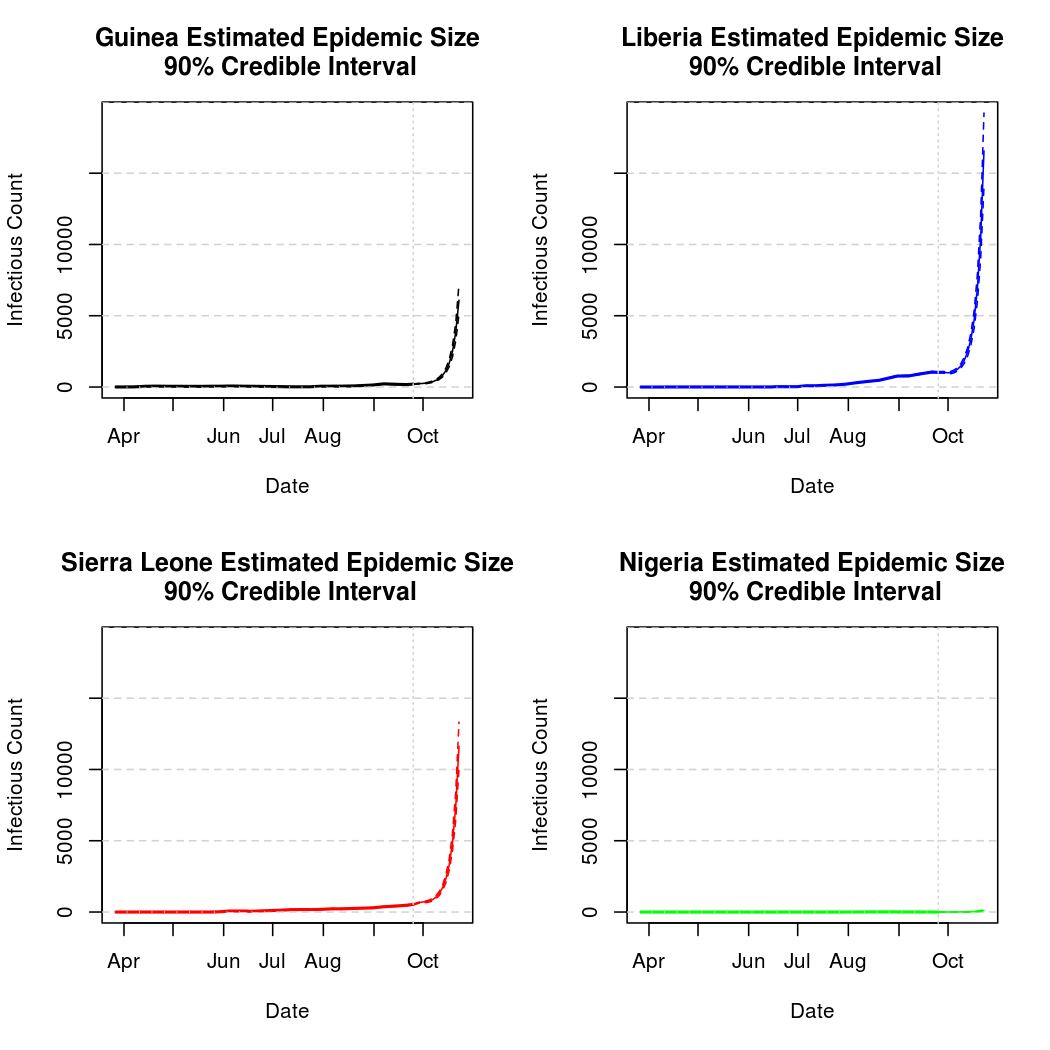
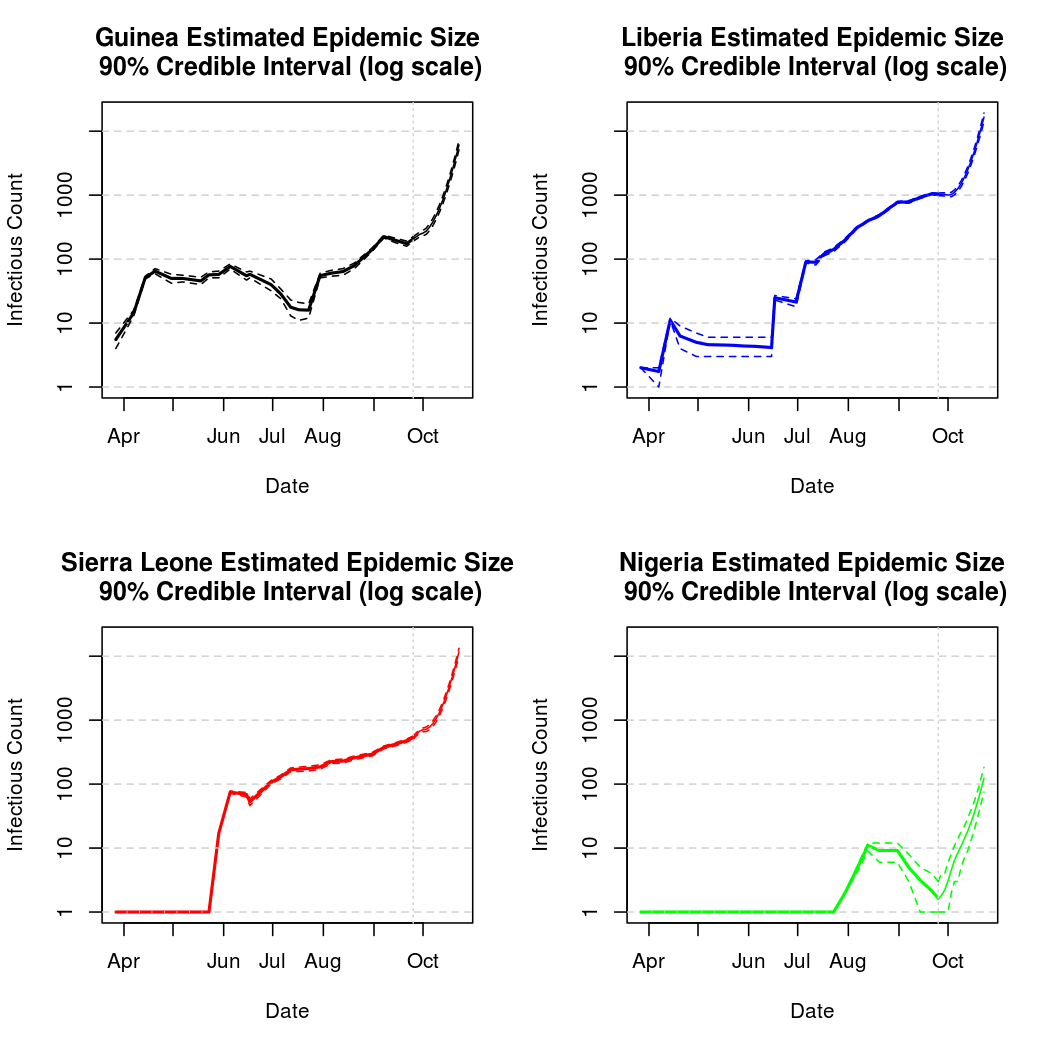
It can also be helpful to take a look at the data in tabular form.
Estimated and Predicted number of Infectious Individuals
| Guinea | Liberia | Sierra Leone | Nigeria | |
|---|---|---|---|---|
| 2014-03-27 | 5 | 1 | 0 | 0 |
| 2014-04-14 | 50 | 10 | 0 | 0 |
| 2014-04-30 | 49 | 4 | 0 | 0 |
| 2014-05-18 | 45 | 4 | 0 | 0 |
| 2014-05-29 | 57 | 3 | 16 | 0 |
| 2014-06-15 | 54 | 3 | 68 | 0 |
| 2014-06-30 | 39 | 20 | 105 | 0 |
| 2014-07-12 | 17 | 88 | 166 | 0 |
| 2014-07-23 | 15 | 141 | 174 | 0 |
| 2014-08-06 | 59 | 305 | 224 | 4 |
| 2014-08-20 | 78 | 479 | 257 | 8 |
| 2014-09-07 | 224 | 784 | 374 | 4 |
| 2014-09-21 | 173 | 1050 | 477 | 1 |
| 2014-09-25 | 207 | 1026 | 536 | 1 |
| 2014-09-27 | 224 | 1031 | 601 | 1 |
| 2014-09-29 | 243 | 1027 | 674 | 1 |
| 2014-10-01 | 254 | 1001 | 707 | 2 |
| 2014-10-03 | 273 | 1023 | 722 | 4 |
| 2014-10-05 | 316 | 1110 | 778 | 5 |
| 2014-10-07 | 379 | 1263 | 882 | 7 |
| 2014-10-09 | 474 | 1507 | 1050 | 10 |
| 2014-10-11 | 614 | 1888 | 1312 | 13 |
| 2014-10-13 | 827 | 2461 | 1712 | 17 |
| 2014-10-15 | 1153 | 3352 | 2331 | 24 |
| 2014-10-17 | 1665 | 4735 | 3300 | 34 |
| 2014-10-19 | 2484 | 6955 | 4853 | 51 |
| 2014-10-21 | 3831 | 10552 | 7383 | 78 |
| 2014-10-23 | 6089 | 16565 | 11618 | 122 |
Such data can also be visualized in map form, though the recent surge in predicted cases has the effect of swamping the earlier dynamics:
Total Infection Size - Estimated and Predicted:
Conclusions
This epidemic is evolving extremely rapidly. As of 8/12, it looked like the situation in Liberia was set to continue worsening, and that Guinea is at risk of the same (though not to nearly the same degree). On the other hand, the epidemic in Sierra Leone appeared to be leveling off (though not disappearing). As of 8/28, these look like reasonable predictions, however the models appear to have resumed predicting a fairly catastrophic continued spread, especially in liberia. In particular, the models predict that the epidemic will take off in Nigeria, as the countries are assumed in this case to share several intensity parameters. We may hope that this particular simplifying assumption is invalid, however it is not a hopeful sign that WHO predictions are also becoming catastrophic. These models can not anticipate public health interventions and sudden changes in governmental policy and individual behavior, but recent news from the region gives little reason to hope for a swift end to the epidemic. It is more important now than ever to support the efforts of involved governmental and non-governmental organizations like the WHO and MSF
That wraps up the analyses for now. This document will continue to be updated as the epidemic progresses, reflecting new data and perhaps additional analysis techniques. As the document is tracked via source control it will be easy to see how well past predictions held up and how they change in response to new information. Questions and comments can be shared here